|
|
2 years ago | |
|---|---|---|
| .github | 2 years ago | |
| colossalai | 2 years ago | |
| docker | 2 years ago | |
| docs | 2 years ago | |
| examples | 2 years ago | |
| inference@7e3dd8c27e | 2 years ago | |
| requirements | 2 years ago | |
| tests | 2 years ago | |
| .clang-format | ||
| .flake8 | ||
| .gitignore | 2 years ago | |
| .gitmodules | ||
| .isort.cfg | 2 years ago | |
| .pre-commit-config.yaml | 2 years ago | |
| .readthedocs.yaml | 2 years ago | |
| .style.yapf | ||
| CHANGE_LOG.md | ||
| CONTRIBUTING.md | ||
| LICENSE | 2 years ago | |
| MANIFEST.in | 2 years ago | |
| README-zh-Hans.md | 2 years ago | |
| README.md | 2 years ago | |
| pytest.ini | ||
| setup.py | 2 years ago | |
| version.txt | 2 years ago | |
README.md
Colossal-AI
Colossal-AI: A Unified Deep Learning System for Big Model Era
Paper | Documentation | Examples | Forum | Blog
Table of Contents
- Why Colossal-AI
- Features
- Parallel Training Demo
- Single GPU Training Demo
- Inference (Energon-AI) Demo
- Colossal-AI for Real World Applications
- Installation
- Use Docker
- Community
- Contributing
- Cite Us
Why Colossal-AI

Prof. James Demmel (UC Berkeley): Colossal-AI makes training AI models efficient, easy, and scalable.
Features
Colossal-AI provides a collection of parallel components for you. We aim to support you to write your distributed deep learning models just like how you write your model on your laptop. We provide user-friendly tools to kickstart distributed training and inference in a few lines.
-
Parallelism strategies
- Data Parallelism
- Pipeline Parallelism
- 1D, 2D, 2.5D, 3D Tensor Parallelism
- Sequence Parallelism
- Zero Redundancy Optimizer (ZeRO)
-
Heterogeneous Memory Management
-
Friendly Usage
- Parallelism based on configuration file
-
Inference
-
Colossal-AI in the Real World
- Biomedicine: FastFold accelerates training and inference of AlphaFold protein structure
Parallel Training Demo
ViT
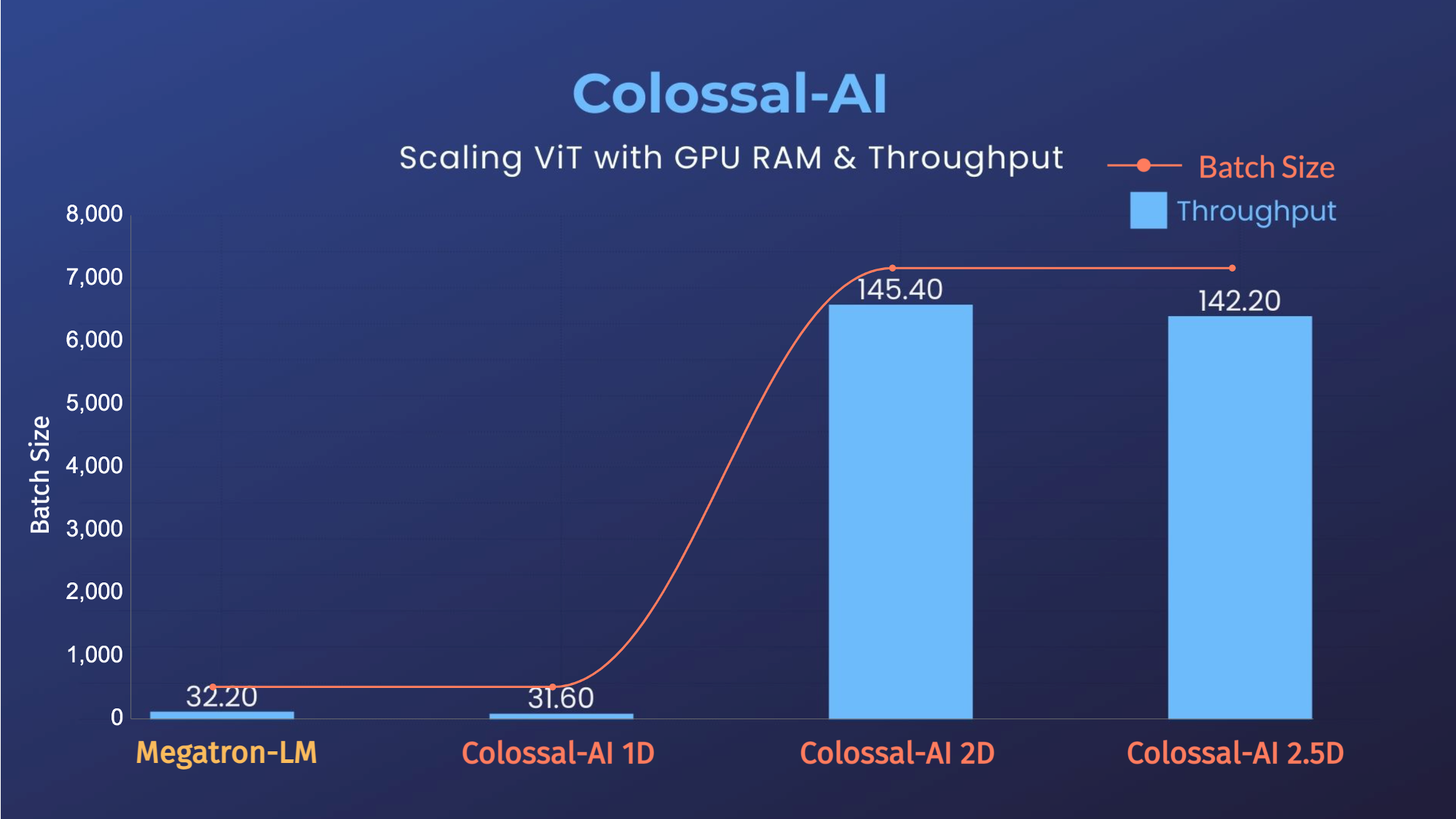
- 14x larger batch size, and 5x faster training for Tensor Parallelism = 64
GPT-3
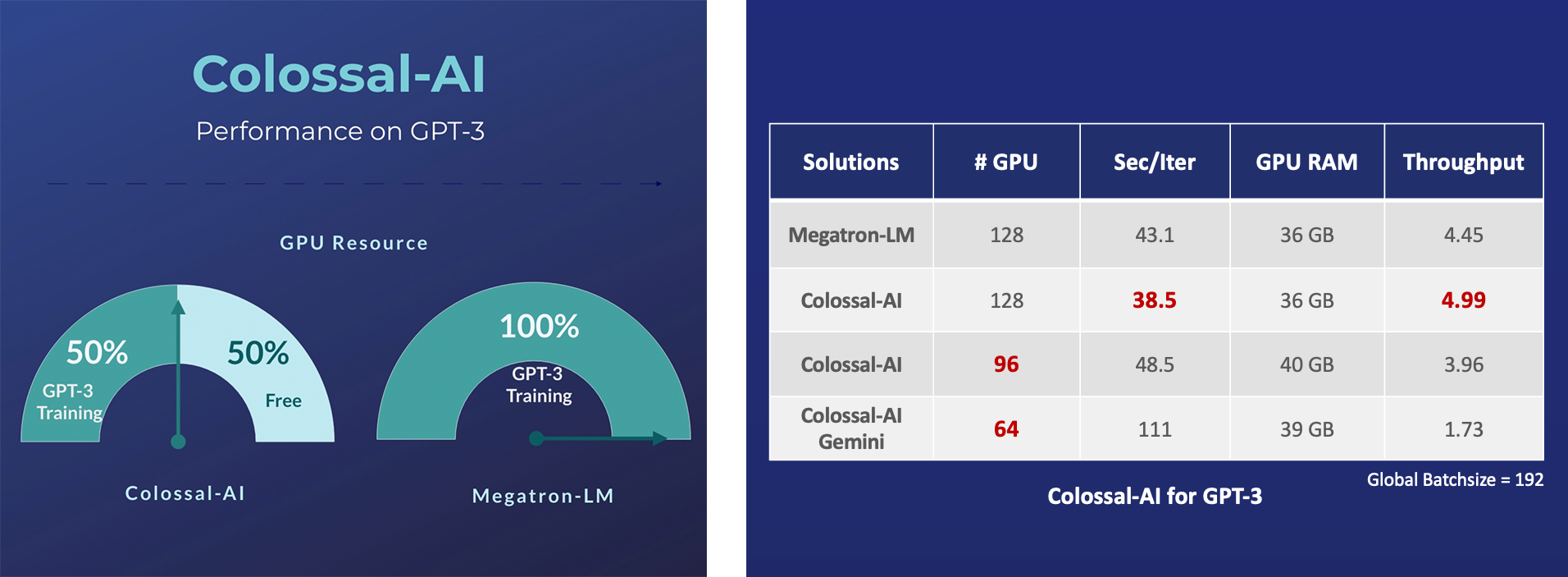
- Save 50% GPU resources, and 10.7% acceleration
GPT-2
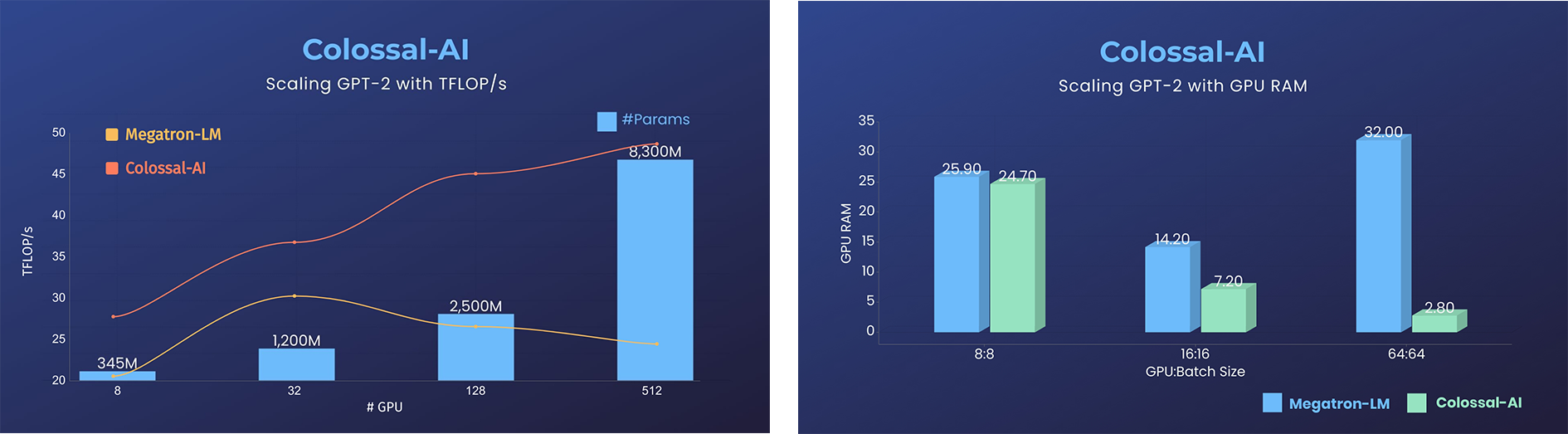
- 11x lower GPU memory consumption, and superlinear scaling efficiency with Tensor Parallelism
GPT-2.png)
- 24x larger model size on the same hardware
- over 3x acceleration
BERT
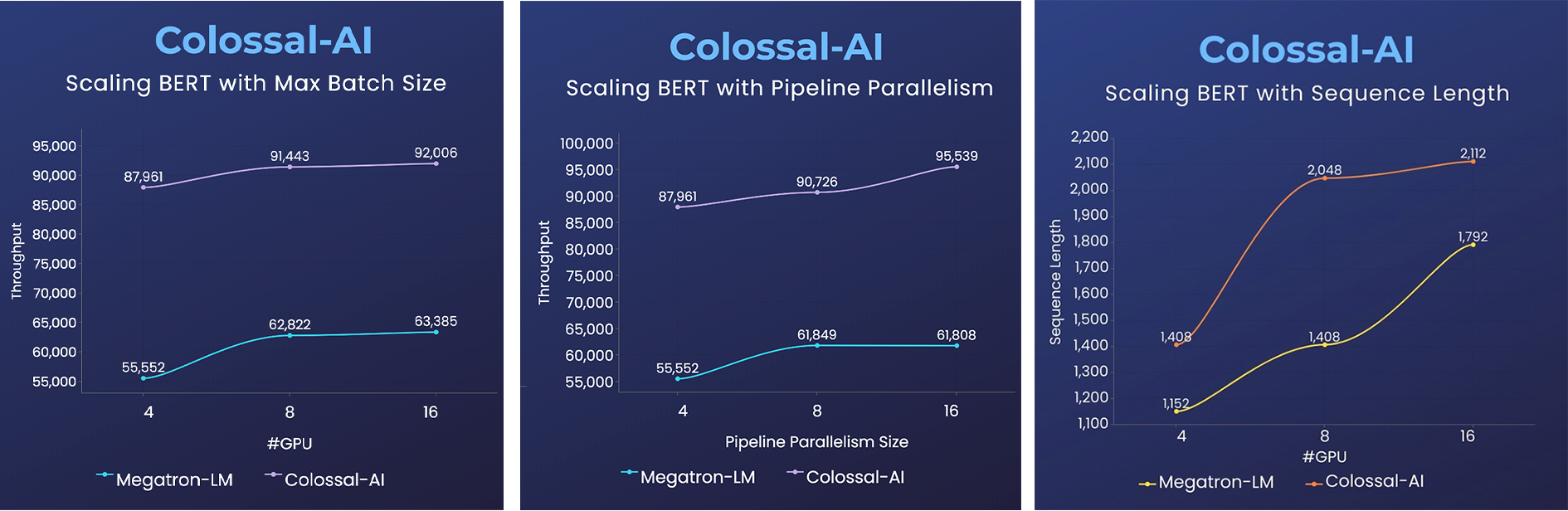
- 2x faster training, or 50% longer sequence length
PaLM
- PaLM-colossalai: Scalable implementation of Google's Pathways Language Model (PaLM).
OPT
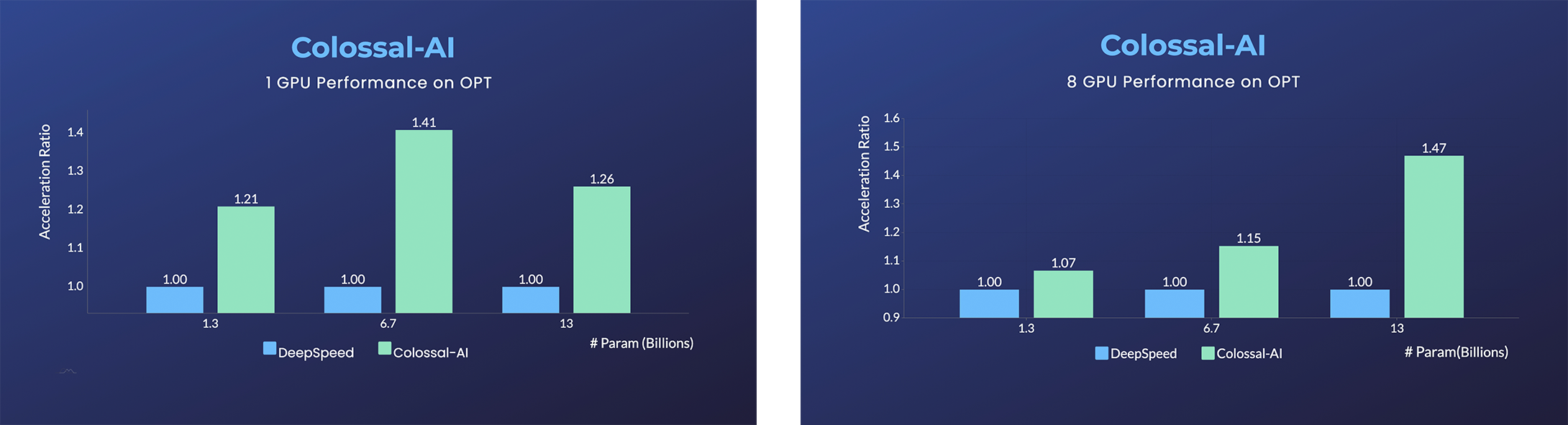
- Open Pretrained Transformer (OPT), a 175-Billion parameter AI language model released by Meta, which stimulates AI programmers to perform various downstream tasks and application deployments because public pretrained model weights.
- 45% speedup fine-tuning OPT at low cost in lines. [Example] [Online Serving]
Please visit our documentation and examples for more details.
Recommendation System Models
- Cached Embedding, utilize software cache to train larger embedding tables with a smaller GPU memory budget.
Single GPU Training Demo
GPT-2
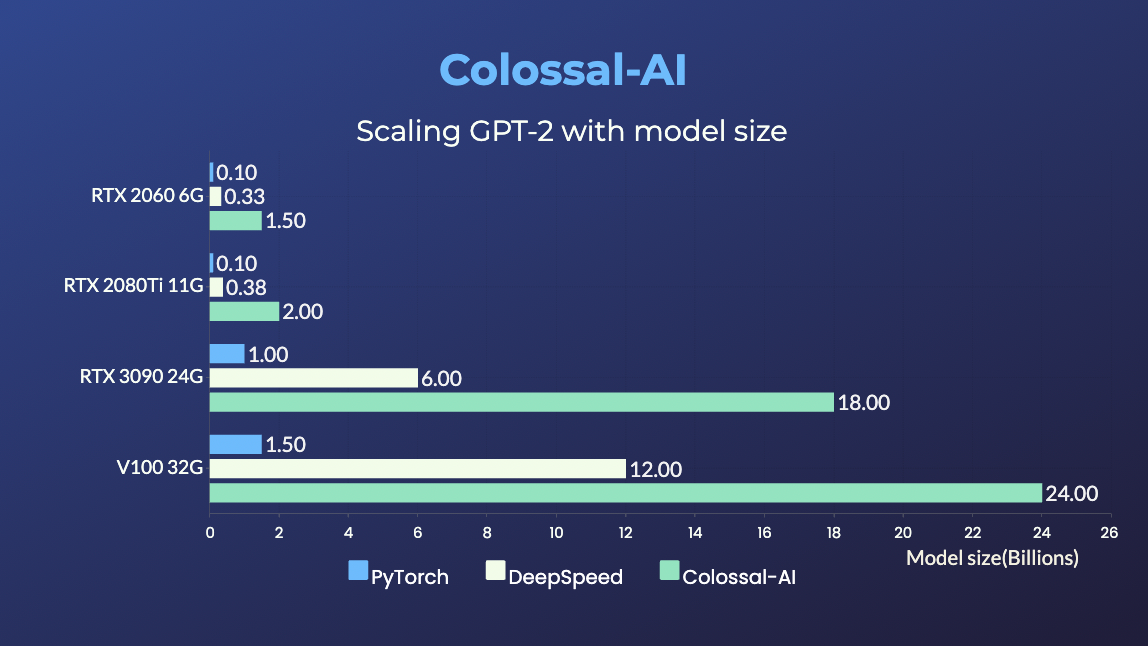
- 20x larger model size on the same hardware
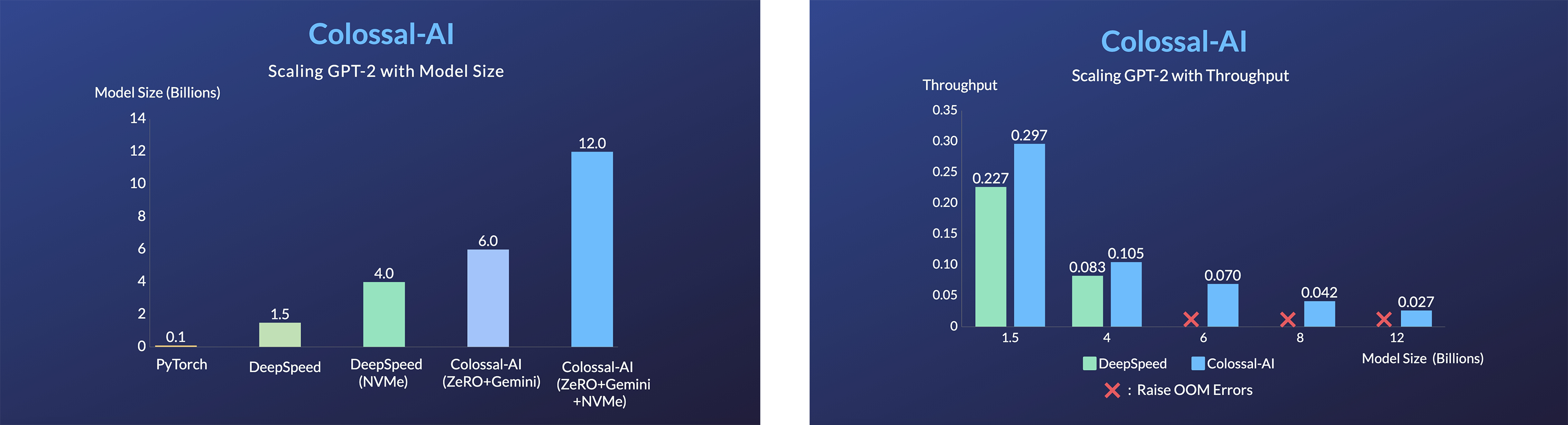
- 120x larger model size on the same hardware (RTX 3080)
PaLM
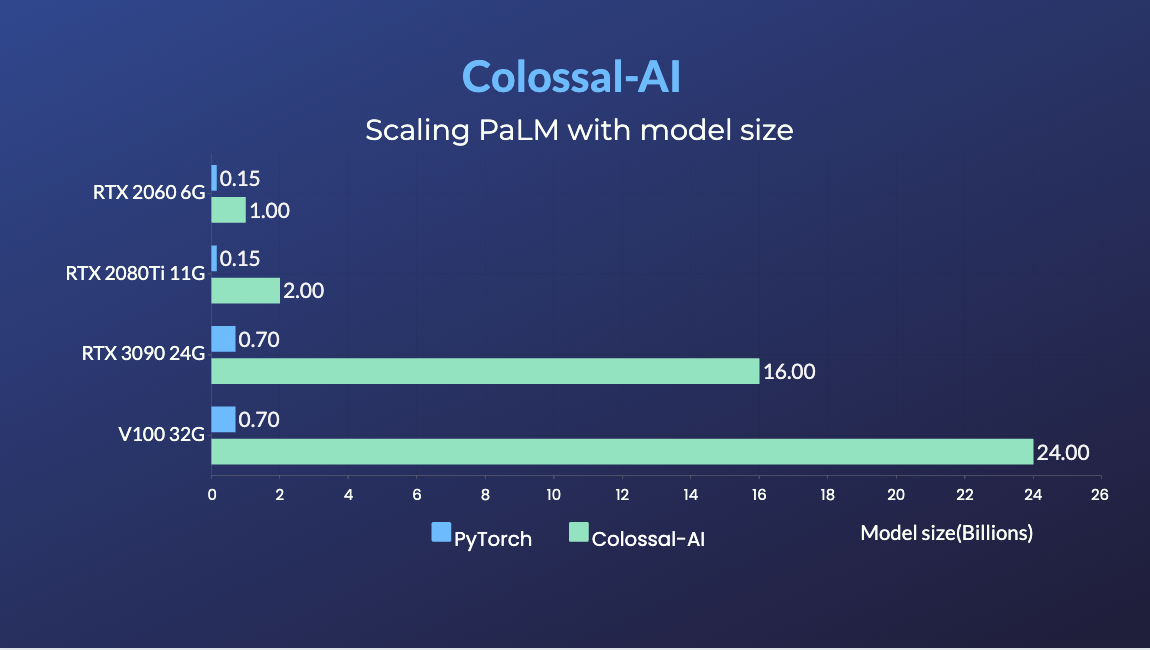
- 34x larger model size on the same hardware
Inference (Energon-AI) Demo
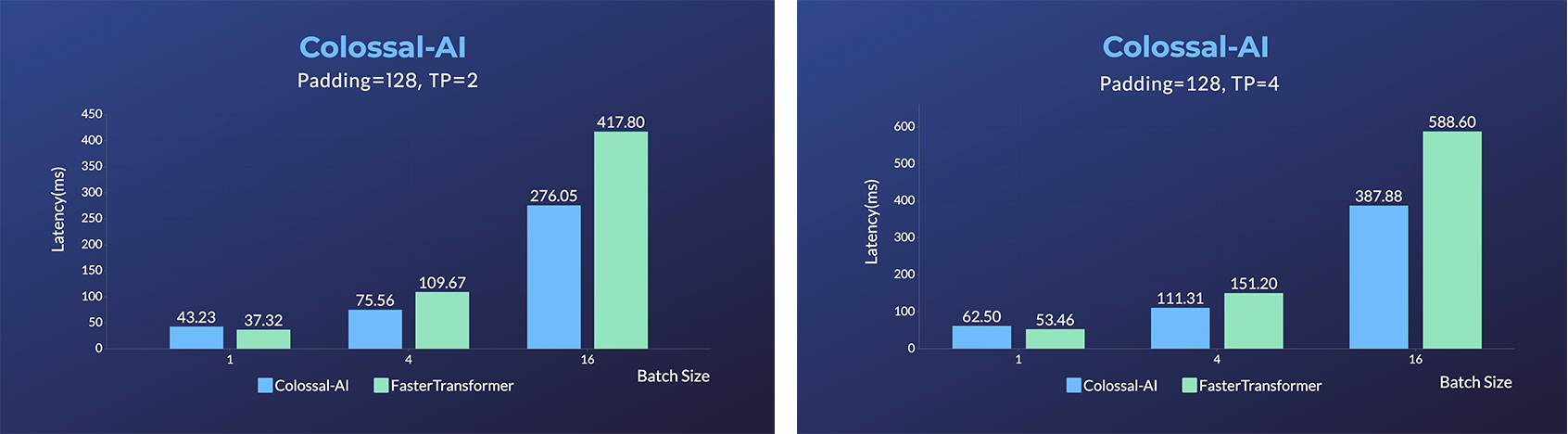
- Energon-AI: 50% inference acceleration on the same hardware

- OPT Serving: Try 175-billion-parameter OPT online services for free, without any registration whatsoever.
Colossal-AI in the Real World
AIGC
Acceleration of AIGC (AI-Generated Content) models such as Stable Diffusion
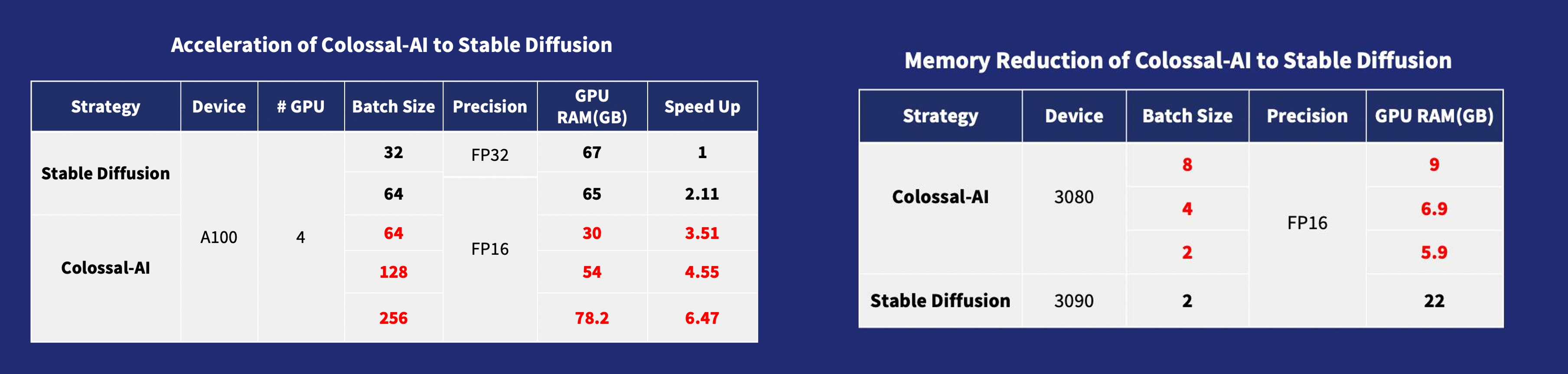
- Stable Diffusion with Colossal-AI: 6.5x faster training and pretraining cost saving, the hardware cost of fine-tuning can be almost 7X cheaper (from RTX3090/4090 to RTX3050/2070)

Biomedicine
Acceleration of AlphaFold Protein Structure

- FastFold: accelerating training and inference on GPU Clusters, faster data processing, inference sequence containing more than 10000 residues.
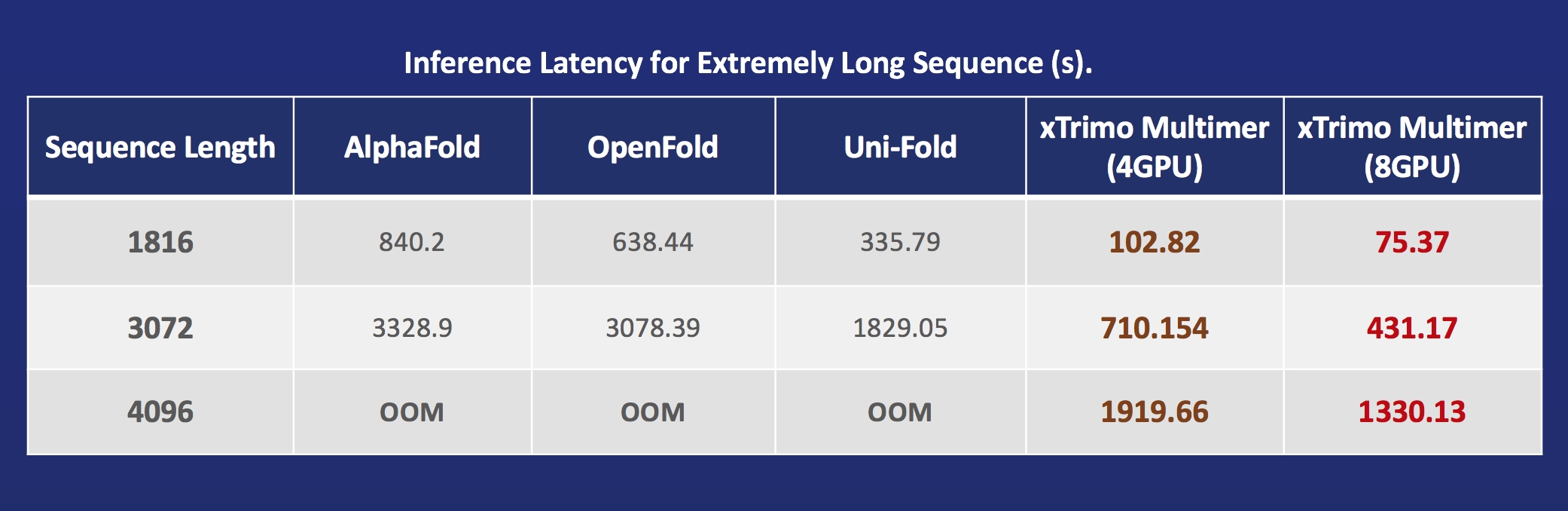
- xTrimoMultimer: accelerating structure prediction of protein monomers and multimer by 11x.
Installation
Download From Official Releases
You can visit the Download page to download Colossal-AI with pre-built CUDA extensions.
Download From Source
The version of Colossal-AI will be in line with the main branch of the repository. Feel free to raise an issue if you encounter any problem. :)
git clone https://github.com/hpcaitech/ColossalAI.git
cd ColossalAI
# install dependency
pip install -r requirements/requirements.txt
# install colossalai
pip install .
If you don't want to install and enable CUDA kernel fusion (compulsory installation when using fused optimizer):
NO_CUDA_EXT=1 pip install .
Use Docker
Pull from DockerHub
You can directly pull the docker image from our DockerHub page. The image is automatically uploaded upon release.
Build On Your Own
Run the following command to build a docker image from Dockerfile provided.
Building Colossal-AI from scratch requires GPU support, you need to use Nvidia Docker Runtime as the default when doing
docker build. More details can be found here. We recommend you install Colossal-AI from our project page directly.
cd ColossalAI
docker build -t colossalai ./docker
Run the following command to start the docker container in interactive mode.
docker run -ti --gpus all --rm --ipc=host colossalai bash
Community
Join the Colossal-AI community on Forum, Slack, and WeChat to share your suggestions, feedback, and questions with our engineering team.
Contributing
If you wish to contribute to this project, please follow the guideline in Contributing.
Thanks so much to all of our amazing contributors!
The order of contributor avatars is randomly shuffled.
Cite Us
@article{bian2021colossal,
title={Colossal-AI: A Unified Deep Learning System For Large-Scale Parallel Training},
author={Bian, Zhengda and Liu, Hongxin and Wang, Boxiang and Huang, Haichen and Li, Yongbin and Wang, Chuanrui and Cui, Fan and You, Yang},
journal={arXiv preprint arXiv:2110.14883},
year={2021}
}



